Automatic Regression Modeling
Source:vignettes/Automatic_Regression_Modeling.Rmd
Automatic_Regression_Modeling.RmdInstallation
You can install autoReg package on github.
#install.packages("devtools")
devtools::install_github("cardiomoon/autoReg")Load package
To load the package, use library() function.
library(autoReg)
library(dplyr)
Attaching package: 'dplyr'
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionLinear model with multiple variables
The package autoReg aims automatic selection of
explanatory variables of regression models. Let’s begin with famous
mtcars data. We select mpg(miles per gallon) as a dependent variable and
select wt(weight), hp(horse power) and am(transmission, 0=automatic,
1=manual) as explanatory variables and included all possible
interaction. The autoReg() function make a table summarizing the result
of analysis.
fit=lm(mpg~wt*hp*am,data=mtcars)
autoReg(fit)
—————————————————————————————————————————————————————————————————————————————————————
Dependent: mpg unit value Coefficient (multivariable)
—————————————————————————————————————————————————————————————————————————————————————
wt [1.5,5.4] Mean ± SD 3.2 ± 1.0 -4.80 (-13.06 to 3.46, p=.242)
hp [52,335] Mean ± SD 146.7 ± 68.6 -0.09 (-0.22 to 0.04, p=.183)
am [0,1] Mean ± SD 0.4 ± 0.5 12.84 (-16.52 to 42.19, p=.376)
wt:hp 0.01 (-0.03 to 0.05, p=.458)
wt:am -5.36 (-14.85 to 4.13, p=.255)
hp:am -0.03 (-0.22 to 0.15, p=.717)
wt:hp:am : 0.02 (-0.04 to 0.07, p=.503)
—————————————————————————————————————————————————————————————————————————————————————You can make a publication-ready table easily using myft(). It makes a flextable object which can use in HTML, PDF, microsoft word or powerpoint file.
Dependent: mpg |
|
unit |
value |
Coefficient (multivariable) |
|---|---|---|---|---|
wt |
[1.5,5.4] |
Mean ± SD |
3.2 ± 1.0 |
-4.80 (-13.06 to 3.46, p=.242) |
hp |
[52,335] |
Mean ± SD |
146.7 ± 68.6 |
-0.09 (-0.22 to 0.04, p=.183) |
am |
[0,1] |
Mean ± SD |
0.4 ± 0.5 |
12.84 (-16.52 to 42.19, p=.376) |
wt:hp |
0.01 (-0.03 to 0.05, p=.458) |
|||
wt:am |
-5.36 (-14.85 to 4.13, p=.255) |
|||
hp:am |
-0.03 (-0.22 to 0.15, p=.717) |
|||
wt:hp:am |
: |
0.02 (-0.04 to 0.07, p=.503) |
||
From the result of multivariable analysis, we found no explanatory variable is significant.
Selection of explanatory variable from univariable model
You can start with univariable model. With a list of univariable model, you can select potentially significant explanatory variable(p value below 0.2 for example). The autoReg() function automatically select from univariable model with a given p value threshold(default value is 0.2).
Dependent: mpg |
|
unit |
value |
Coefficient (univariable) |
Coefficient (multivariable) |
|---|---|---|---|---|---|
wt |
[1.5,5.4] |
Mean ± SD |
3.2 ± 1.0 |
-5.34 (-6.49 to -4.20, p<.001) |
-7.50 (-13.21 to -1.80, p=.012) |
hp |
[52,335] |
Mean ± SD |
146.7 ± 68.6 |
-0.07 (-0.09 to -0.05, p<.001) |
-0.11 (-0.20 to -0.02, p=.022) |
am |
[0,1] |
Mean ± SD |
0.4 ± 0.5 |
7.24 (3.64 to 10.85, p<.001) |
1.91 (-11.29 to 15.10, p=.769) |
wt:hp |
-0.01 (-0.02 to -0.01, p<.001) |
0.02 (-0.00 to 0.05, p=.072) |
|||
wt:am |
1.89 (0.25 to 3.52, p=.025) |
-0.60 (-4.92 to 3.73, p=.778) |
|||
hp:am |
0.01 (-0.02 to 0.04, p=.452) |
||||
wt:hp:am |
: |
-0.00 (-0.01 to 0.01, p=.982) |
|||
As you can see in the above table, the coefficients of hp:am(the interaction of hp and am) and wt:hp:am (interaction of wt and hp and am) have p-values above 0.2. So these variables are excluded and the remaining variables are used in multivariable model. If you want to use all the explanatory variables in the multivariable model, set the threshold 1.
Stepwise backward elimination
From the multivariable model, you can perform stepwise backward elimination with step() function.
fit=lm(mpg~hp+wt+am+wt:hp+wt:am,data=mtcars)
final=step(fit,trace=0)
summary(final)
Call:
lm(formula = mpg ~ hp + wt + hp:wt, data = mtcars)
Residuals:
Min 1Q Median 3Q Max
-3.0632 -1.6491 -0.7362 1.4211 4.5513
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 49.80842 3.60516 13.816 5.01e-14 ***
hp -0.12010 0.02470 -4.863 4.04e-05 ***
wt -8.21662 1.26971 -6.471 5.20e-07 ***
hp:wt 0.02785 0.00742 3.753 0.000811 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.153 on 28 degrees of freedom
Multiple R-squared: 0.8848, Adjusted R-squared: 0.8724
F-statistic: 71.66 on 3 and 28 DF, p-value: 2.981e-13You can perform univariable analysis for variable selection, multivariable analysis and stepwise backward elimination in one step.
Dependent: mpg |
|
unit |
value |
Coefficient (univariable) |
Coefficient (multivariable) |
Coefficient (final) |
|---|---|---|---|---|---|---|
hp |
[52,335] |
Mean ± SD |
146.7 ± 68.6 |
-0.07 (-0.09 to -0.05, p<.001) |
-0.11 (-0.20 to -0.02, p=.022) |
-0.12 (-0.17 to -0.07, p<.001) |
wt |
[1.5,5.4] |
Mean ± SD |
3.2 ± 1.0 |
-5.34 (-6.49 to -4.20, p<.001) |
-7.50 (-13.21 to -1.80, p=.012) |
-8.22 (-10.82 to -5.62, p<.001) |
am |
[0,1] |
Mean ± SD |
0.4 ± 0.5 |
7.24 (3.64 to 10.85, p<.001) |
1.91 (-11.29 to 15.10, p=.769) |
|
hp:wt |
-0.01 (-0.02 to -0.01, p<.001) |
0.02 (-0.00 to 0.05, p=.072) |
0.03 (0.01 to 0.04, p<.001) |
|||
hp:am |
0.01 (-0.02 to 0.04, p=.452) |
|||||
wt:am |
1.89 (0.25 to 3.52, p=.025) |
-0.60 (-4.92 to 3.73, p=.778) |
||||
hp:wt:am |
: |
-0.00 (-0.01 to 0.01, p=.982) |
||||
Linear model with interaction between categorical variable
You can use autoReg() function for models with interaction with categorical variable(s).
Dependent: Sepal.Length |
|
unit |
value |
Coefficient (univariable) |
Coefficient (multivariable) |
Coefficient (final) |
|---|---|---|---|---|---|---|
Sepal.Width |
[2,4.4] |
Mean ± SD |
3.1 ± 0.4 |
-0.22 (-0.53 to 0.08, p=.152) |
0.69 (0.36 to 1.02, p<.001) |
0.80 (0.59 to 1.01, p<.001) |
Species |
setosa (N=50) |
Mean ± SD |
5.0 ± 0.4 |
|||
versicolor (N=50) |
Mean ± SD |
5.9 ± 0.5 |
0.93 (0.73 to 1.13, p<.001) |
0.90 (-0.68 to 2.48, p=.261) |
1.46 (1.24 to 1.68, p<.001) |
|
virginica (N=50) |
Mean ± SD |
6.6 ± 0.6 |
1.58 (1.38 to 1.79, p<.001) |
1.27 (-0.35 to 2.88, p=.123) |
1.95 (1.75 to 2.14, p<.001) |
|
Sepal.Width:Species |
setosa |
0.48 (0.29 to 0.67, p<.001) |
||||
Sepal.Width:Species |
versicolor |
0.93 (0.69 to 1.17, p<.001) |
0.17 (-0.34 to 0.69, p=.503) |
|||
Sepal.Width:Species |
virginica |
1.08 (0.86 to 1.30, p<.001) |
0.21 (-0.29 to 0.72, p=.411) |
|||
Missing data - automatic multiple imputation
Original data
Let us think about linear regression model with iris data. In this model, Sepal.Length is the dependent variable and Sepal.Width and Species are explanatory variables. You can make a table summarizing the result as follows.
name |
levels |
unit |
value |
|---|---|---|---|
Sepal.Width |
[2,4.4] |
Mean ± SD |
3.1 ± 0.4 |
Species |
setosa (N=50) |
Mean ± SD |
5.0 ± 0.4 |
versicolor (N=50) |
Mean ± SD |
5.9 ± 0.5 |
|
virginica (N=50) |
Mean ± SD |
6.6 ± 0.6 |
fit=lm(Sepal.Length~Sepal.Width+Species,data=iris)
df=addFitSummary(df,fit,"Coefficient (original data)")
df %>% myft()name |
levels |
unit |
value |
Coefficient (original data) |
|---|---|---|---|---|
Sepal.Width |
[2,4.4] |
Mean ± SD |
3.1 ± 0.4 |
0.80 (0.59 to 1.01, p<.001) |
Species |
setosa (N=50) |
Mean ± SD |
5.0 ± 0.4 |
|
versicolor (N=50) |
Mean ± SD |
5.9 ± 0.5 |
1.46 (1.24 to 1.68, p<.001) |
|
virginica (N=50) |
Mean ± SD |
6.6 ± 0.6 |
1.95 (1.75 to 2.14, p<.001) |
Missed data
For simulation of the MCAR(missing at completely random) data, one third of the Sepal.Width records are replace with NA(missing). If you want to perform missing data analysis, use gaze() function with missing=TRUE.
iris1=iris
set.seed=123
no=sample(1:150,50,replace=FALSE)
iris1$Sepal.Width[no]=NA
gaze(Sepal.Width~.,data=iris1,missing=TRUE) %>% myft()Dependent:Sepal.Width |
levels |
Not missing (N=100) |
Missing (N=50) |
p |
|---|---|---|---|---|
Sepal.Length |
Mean ± SD |
5.8 ± 0.8 |
5.9 ± 0.8 |
.304 |
Petal.Length |
Mean ± SD |
3.7 ± 1.8 |
3.9 ± 1.7 |
.368 |
Petal.Width |
Mean ± SD |
1.2 ± 0.8 |
1.2 ± 0.7 |
.613 |
Species |
setosa |
35 (35%) |
15 (30%) |
.811 |
versicolor |
32 (32%) |
18 (36%) |
||
virginica |
33 (33%) |
17 (34%) |
And then we do same analysis with this data.
fit1=lm(Sepal.Length~Sepal.Width+Species,data=iris1)
df=addFitSummary(df,fit1,"Coefficient (missed data)")
df %>% myft()name |
levels |
unit |
value |
Coefficient (original data) |
Coefficient (missed data) |
|---|---|---|---|---|---|
Sepal.Width |
[2,4.4] |
Mean ± SD |
3.1 ± 0.4 |
0.80 (0.59 to 1.01, p<.001) |
1.04 (0.74 to 1.34, p<.001) |
Species |
setosa (N=50) |
Mean ± SD |
5.0 ± 0.4 |
||
versicolor (N=50) |
Mean ± SD |
5.9 ± 0.5 |
1.46 (1.24 to 1.68, p<.001) |
1.55 (1.26 to 1.84, p<.001) |
|
virginica (N=50) |
Mean ± SD |
6.6 ± 0.6 |
1.95 (1.75 to 2.14, p<.001) |
2.13 (1.86 to 2.40, p<.001) |
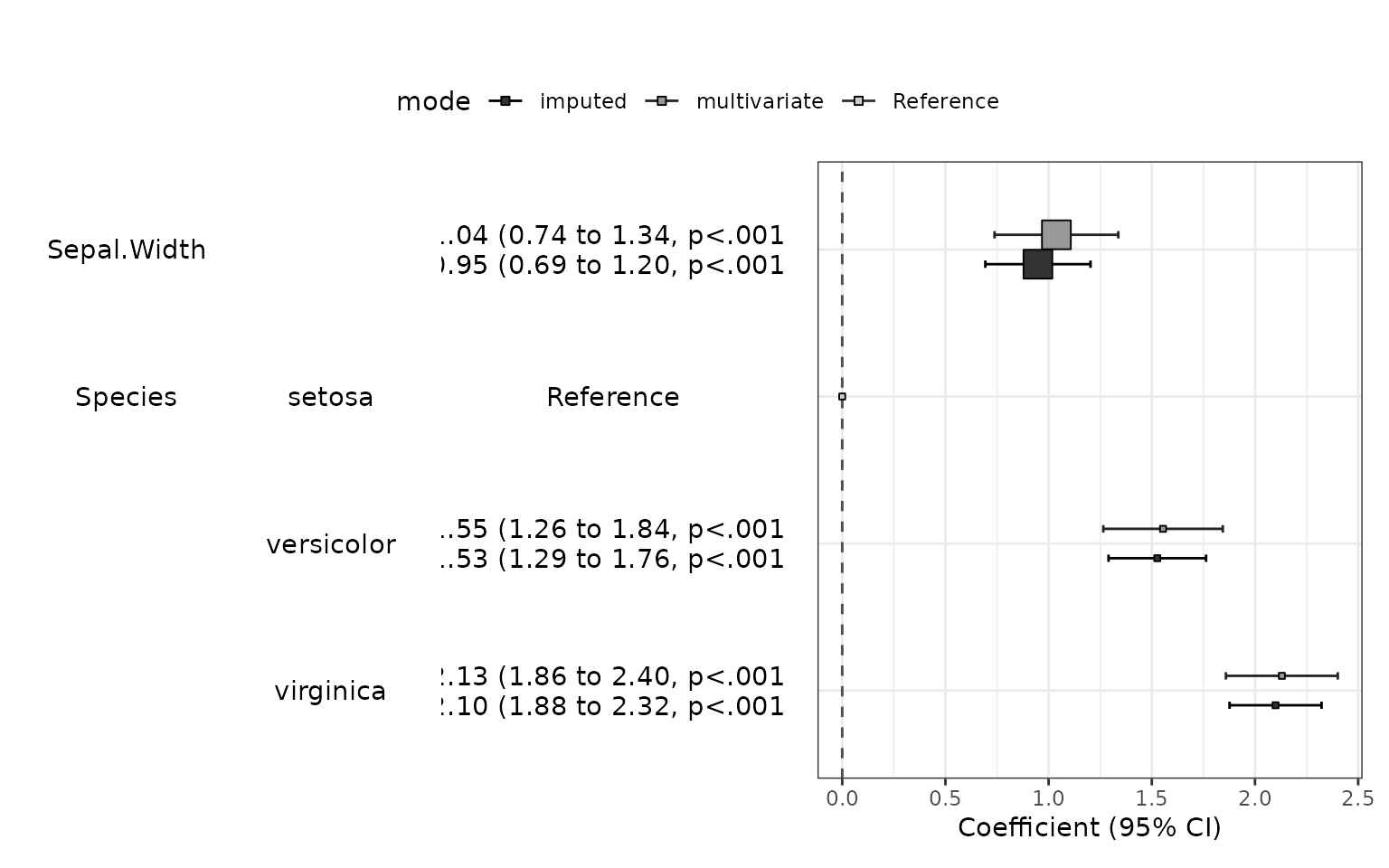
Multiple imputation
You can do multiple imputation by using imputedReg() function. This function perform multiple imputation using mice() function in mice package. The default value of the number of multiple imputation is 20. You can adjust the number with m argument. You can set random number generator with seed argument.
fit2=imputedReg(fit1, m=20,seed=1234)
df=addFitSummary(df,fit2,statsname="Coefficient (imputed)")
df %>% myft()name |
levels |
unit |
value |
Coefficient (original data) |
Coefficient (missed data) |
Coefficient (imputed) |
|---|---|---|---|---|---|---|
Sepal.Width |
[2,4.4] |
Mean ± SD |
3.1 ± 0.4 |
0.80 (0.59 to 1.01, p<.001) |
1.04 (0.74 to 1.34, p<.001) |
0.95 (0.69 to 1.20, p<.001) |
Species |
setosa (N=50) |
Mean ± SD |
5.0 ± 0.4 |
|||
versicolor (N=50) |
Mean ± SD |
5.9 ± 0.5 |
1.46 (1.24 to 1.68, p<.001) |
1.55 (1.26 to 1.84, p<.001) |
1.53 (1.29 to 1.76, p<.001) |
|
virginica (N=50) |
Mean ± SD |
6.6 ± 0.6 |
1.95 (1.75 to 2.14, p<.001) |
2.13 (1.86 to 2.40, p<.001) |
2.10 (1.88 to 2.32, p<.001) |
You can make a plot summarizing models with modelPlot() function.
modelPlot(fit1,imputed=TRUE)